Biosynthesis
Our central focus is on how microorganisms, particularly bacteria actinomycetes, produce specialized metabolites. These molecules are complex, and even organic chemists would struggle to synthesize them in the laboratory. What sets microorganisms apart are their unique enzymes. These enzymes have the remarkable ability to take seemingly uninteresting molecules and transform them, step-by-step, into compounds that are special, complex, and biologically active. We study these enzymes and how they function and biosynthesize specialized metabolites. These studies serve as a prerequisite for our efforts to develop more efficient antibiotics by modifying biosynthesis or conducting genome-level data mining. Moreover, certain enzymes merit individual study owing to their unexpected functions, utilization of unusual cofactors, or novel protein structures. These attributes often reveal novel phenomena within nature.
Biosynthesis of Lincosamides
Lincosamide antibiotics hamper protein synthesis within bacterial cells by binding to the 50S ribosomal subunit. Lincosamides are naturally produced by actinomycetes, forming a small group of natural products with simple structures, composed primarily of a single amino acid and amino thiosugar. However, the assembly of lincomycin, a representative lincosamide, requires more than 20 biosynthetic enzymes, and it involves remarkably complex and intriguing mechanisms. Genetic engineering to prepare knock-out strains, heterologous production and purification of enzymes, enzymatic assays, liquid chromatography with mass spectroscopy, and crystallography are the most prominent techniques that we are using to uncover biosynthetic machineries of bioactive metabolites.


Among the enzymes involved in lincosamide biosynthesis, we were particularly intrigued by oxidoreductases that utilize F420, an unusual deazaflavin cofactor. Our investigation, in collaboration with Ghader Bashiri from the University of Auckland, revealed that within this group of enzymes, some catalyze a common step—reduction of a double bond—while others exhibit a more striking function. Specifically, they catalyze the reduction of a conjugated system of two double bonds—a reaction that is uncommon in nature. Whether one or two double bonds is crucial for the shape of the final metabolites, which, in turn, influences their ability to bind to biological targets. Elucidating the underlying factors behind the differing reaction specificities using structural biology techniques such as crystallography and cryo-EM, are in progress.
Relevant papers:
Typically, enzymes from the TldD/TldE family function as Zn-dependent heterodimeric peptidases or proteases. However, in lincosamide biosynthesis, we discovered the first instance of these proteins catalyzing N-deacetylation. What is more, this step is crucial as it unblocks an amino group of a non-bioactive lincosamide intermediate, enabling its subsequent processing to a bioactive molecule.
Relevant papers
The amino acid of lincomycin closely resembles proteinogenic L-proline, albeit with a three-carbon tail attached to it. Interestingly, this unusual alkyl-L-proline does not derive from L-proline itself, but from L-tyrosine, which undergoes complete restructuring by the strain to resemble L-proline. This transformation necessitates the action of six biosynthetic proteins. In our laboratory, we conducted experiments that provided insights into the intriguing pathway through which this transformation occurs.
Relevant papers

Preparation of more Efficient Antibiotics
Detailed understanding of how actinomycetes biosynthesize bioactive molecules presents a valuable opportunity to develop enhanced drug leads. The sequence of biosynthetic steps, enzyme promiscuity, reaction specificity, and catalytic mechanisms serve as a blueprint for directing biosynthesis towards the synthesis of structural analogs that may exhibit greater efficiency or other improved characteristics.
Discovering distinct reaction specificities of pyridoxal-5-phosphate-dependent enzymes in the biosynthesis of lincomycin and celesticetin lincosamides inspired us to employ genetic engineering techniques such as REDIRECT PCR-targeting and the CRISPR-Cas9 method to combine these two pathways. Briefly, we converted a lincosamide intermediate purified from a knock-out of lincomycin-producing strain with five heterologously produced enzymes from celesticetin biosynthesis. In this way, we prepared a groundbreaking lincosamide chimera, CELIN, which exhibited superior antibiotic efficacy compared to other lincosamides. We are now conducting preclinical trials to find out the potential of CELIN to be applied in clinical practice. Currently, we are collaborating with Santiago Lab, a prominent organic synthesis company, to enhance CELIN’s effectiveness through targeted modifications. Our ultimate goal is to develop lincosamides capable of binding to (di)methylated ribosomes, thereby addressing the critical issue of antibiotic resistance, particularly against MRSA (methicillin-resistant Staphylococcus aureus). MRSA represents one of the most challenging pathogens for which new antibiotic solutions are urgently needed.
Relevant papers:
Relevant international patent:
Lincosamide derivatives, preparation and use thereof as antimicrobial agent
Janata, Jiri; Kamenik, Zdenek; (…); Gazak, Radek
Nov 3 2020 |Official Gazette of the United States Patent and Trademark Office Patents
Genome Mining to Discover New Bioactive Metabolites
A significant portion of current antibiotics is derived from actinomycetes. Originally isolated from soil or other environments, these microorganisms were cultured in laboratory settings, and the antibiotics they produced were discovered through well-established bioassay-guided methods, primarily during the 1950s to the 1970s. However, the intensity of screenings, especially of soil actinomycetes, led to a high rate of rediscovery, making this strategy ineffective over time.
While discovering new bioactive metabolites by isolating actinomycetes from natural sources presents challenges, an alternative approach exists. Exploiting the exponentially growing volume of sequencing data in public databases, which contain information on biosynthetic genes of bioactive metabolites, offers a promising solution.
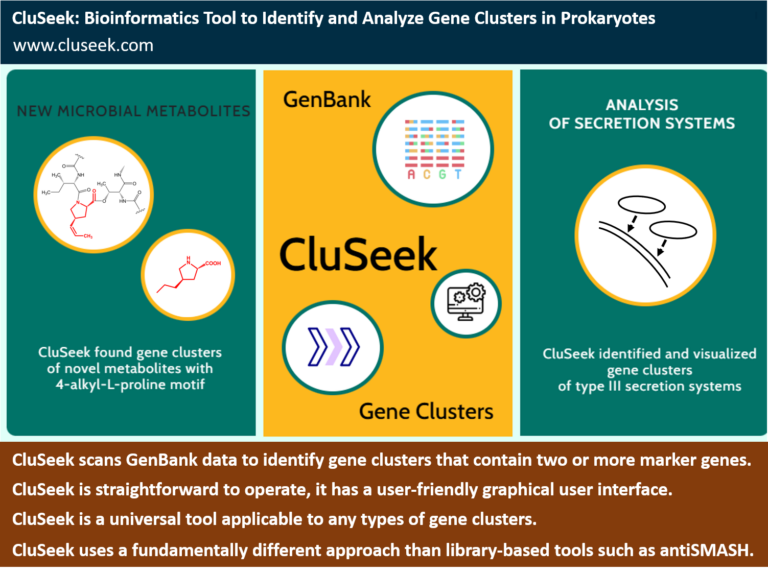
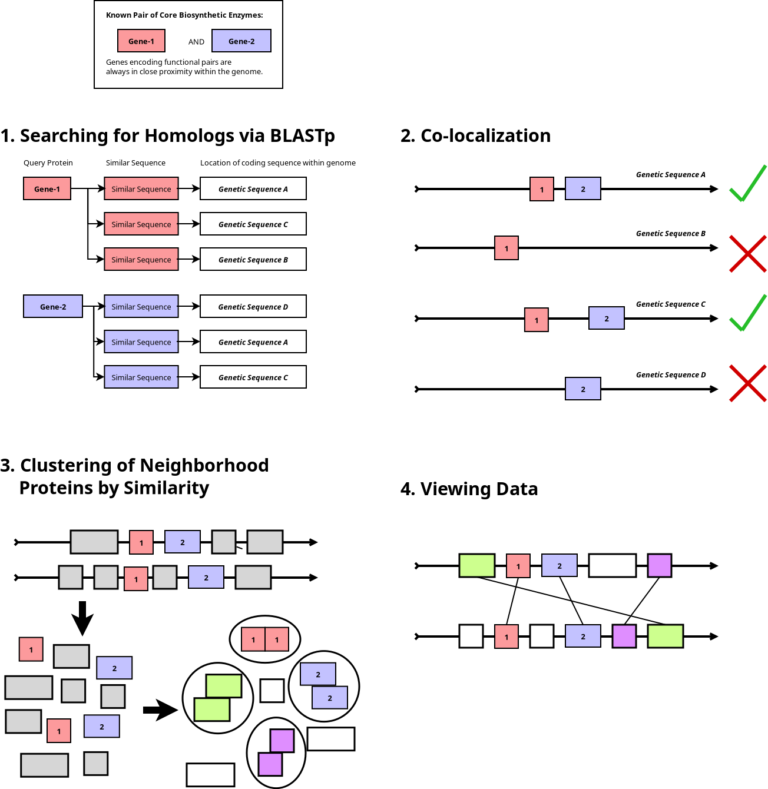
Manual searching through the extensive volume of sequencing data has become nearly impossible today. Motivated by the absence of a suitable bioinformatics tool to automate this process, we have developed one. Named CluSeek, this tool not only identifies gene clusters of interest within the GenBank database but also offers comprehensive analysis. CluSeek is highly versatile, capable of analyzing any type of gene cluster regardless of the encoded phenotype. It is user-friendly and can be easily launched and used by those without IT expertise. CluSeek is publicly accessible so try it out:
We’re intrigued by metabolites with the rare 4-alkyl-L-proline motif due to their increased bioactivity compared to analogs containing the common L-proline. This is true not only for the antibiotic lincomycin (binding) to ribosomes, but also for anticancer pyrrolobenzodiazepines (binding to DNA), some of which share this motif as well. Wondering whether there are more metabolites bearing this valuable motif, we exploited our understanding of how actinomycetes synthesize it. Specifically, we submitted genes involved in 4-alkyl-L-proline biosynthesis to CluSeek, and identified over 200 new gene clusters within GenBank. These correspond to more than 13 structurally distinct classes (compare the three known until now) that presumably contain 4-alkyl-L-proline motif.
Through extensive global collaborations, we now have many of these strains in hands. Our current efforts involve traditional actinomycete culturing, heterologous expression of biosynthetic gene clusters, stable isotope labeling studies, and mass spectrometry metabolomics to pinpoint the actual metabolites. Subsequently, these metabolites will be purified using preparative high-performance liquid chromatography.
Institue of Microbiology
Institute of Microbiology, CAS
Vídeňská 1083, 142 20 Prague 4 - Krč
The Czech Republic
BIOCEV
Institute of Microbiology, CAS
Průmyslová 595, 252 50 Vestec
The Czech Republic
Contact Us
lab111@biomed.cas.cz
+420 241 062 371
