CluSeek
ClusterSeeker
What is CluSeek?
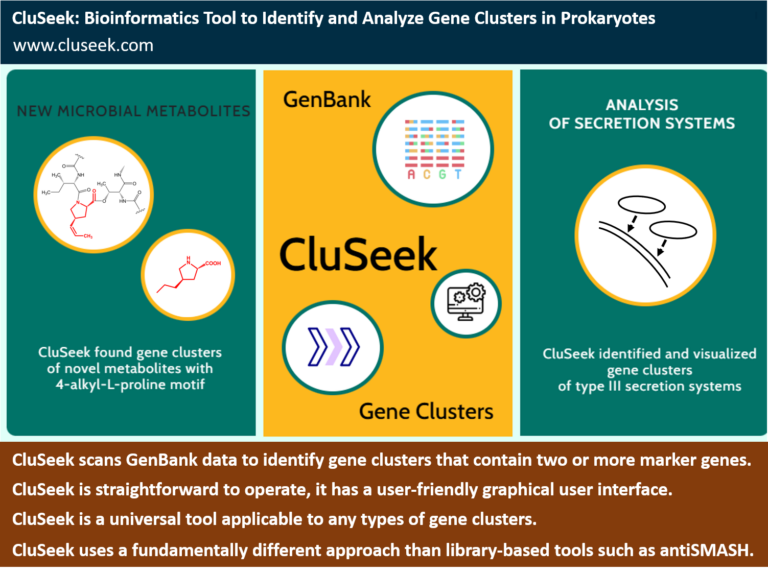
Research of specialized Metabolites:
Is CluSeek similar to antiSMASH?
Is CluSeek easy to use?
Are there any limitations to using CluSeek?
CluSeek has been developed for prokaryotic genomes; however, preliminary testing shows that it can successfully handle also sequencing data from eukaryotes. Currently, CluSeek is fully compatible with Windows, while its adaptation for other platforms is in progress (let us know if it would be relevant to you).
Can I try using CluSeek now?
Please note that the current version of CluSeek remains a work in progress and not all features are fully functional.
Installation:
- Download CluSeek for Windows here , or for MAC here
- and extract the .zip file
- If you are installing CluSeek on Windows for the first time, you will need to install the Windows redistributables (vc_redist.x86) included with CluSeek, or download and install the latest version from microsoft.
- That’s it. Run CluSeek and start a project.
We hope to release additional improvements in the coming weeks.
You can send us your feedback or suggestions at cluseek@biomed.cas.cz
Should you require an older version of CluSeek, please do not hesitate to contact us via email.
Institue of Microbiology
Institute of Microbiology, CAS
Vídeňská 1083, 142 20 Prague 4 - Krč
The Czech Republic
BIOCEV
Institute of Microbiology, CAS
Průmyslová 595, 252 50 Vestec
The Czech Republic
Contact Us
lab111@biomed.cas.cz
+420 241 062 371
